Tidy summarizes information about the components of a model. A model component might be a single term in a regression, a single hypothesis, a cluster, or a class. Exactly what tidy considers to be a model component varies across models but is usually self-evident. If a model has several distinct types of components, you will need to specify which components to return.
Usage
# S3 method for class 'nls'
tidy(x, conf.int = FALSE, conf.level = 0.95, ...)Arguments
- x
An
nlsobject returned fromstats::nls().- conf.int
Logical indicating whether or not to include a confidence interval in the tidied output. Defaults to
FALSE.- conf.level
The confidence level to use for the confidence interval if
conf.int = TRUE. Must be strictly greater than 0 and less than 1. Defaults to 0.95, which corresponds to a 95 percent confidence interval.- ...
Additional arguments. Not used. Needed to match generic signature only. Cautionary note: Misspelled arguments will be absorbed in
..., where they will be ignored. If the misspelled argument has a default value, the default value will be used. For example, if you passconf.lvel = 0.9, all computation will proceed usingconf.level = 0.95. Two exceptions here are:
See also
tidy, stats::nls(), stats::summary.nls()
Other nls tidiers:
augment.nls(),
glance.nls()
Value
A tibble::tibble() with columns:
- conf.high
Upper bound on the confidence interval for the estimate.
- conf.low
Lower bound on the confidence interval for the estimate.
- estimate
The estimated value of the regression term.
- p.value
The two-sided p-value associated with the observed statistic.
- statistic
The value of a T-statistic to use in a hypothesis that the regression term is non-zero.
- std.error
The standard error of the regression term.
- term
The name of the regression term.
Examples
# fit model
n <- nls(mpg ~ k * e^wt, data = mtcars, start = list(k = 1, e = 2))
# summarize model fit with tidiers + visualization
tidy(n)
#> # A tibble: 2 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 k 49.7 3.79 13.1 5.96e-14
#> 2 e 0.746 0.0199 37.5 8.86e-27
augment(n)
#> # A tibble: 32 × 4
#> mpg wt .fitted .resid
#> <dbl> <dbl> <dbl> <dbl>
#> 1 21 2.62 23.0 -2.01
#> 2 21 2.88 21.4 -0.352
#> 3 22.8 2.32 25.1 -2.33
#> 4 21.4 3.22 19.3 2.08
#> 5 18.7 3.44 18.1 0.611
#> 6 18.1 3.46 18.0 0.117
#> 7 14.3 3.57 17.4 -3.11
#> 8 24.4 3.19 19.5 4.93
#> 9 22.8 3.15 19.7 3.10
#> 10 19.2 3.44 18.1 1.11
#> # ℹ 22 more rows
glance(n)
#> # A tibble: 1 × 9
#> sigma isConv finTol logLik AIC BIC deviance df.residual nobs
#> <dbl> <lgl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int>
#> 1 2.67 TRUE 0.00000204 -75.8 158. 162. 214. 30 32
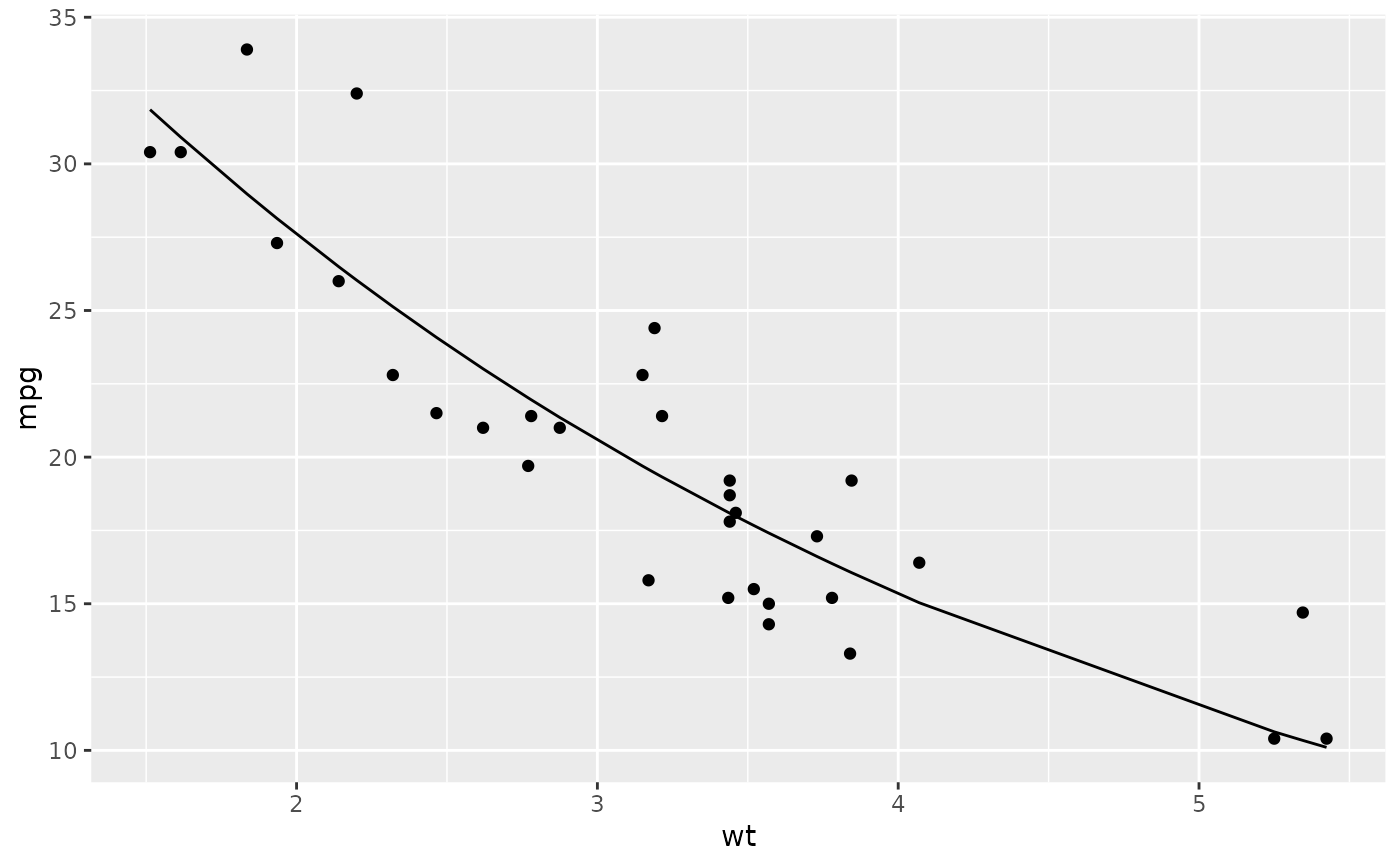
library(ggplot2)
ggplot(augment(n), aes(wt, mpg)) +
geom_point() +
geom_line(aes(y = .fitted))
 newdata <- head(mtcars)
newdata$wt <- newdata$wt + 1
augment(n, newdata = newdata)
#> # A tibble: 6 × 13
#> .rownames mpg cyl disp hp drat wt qsec vs am gear
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Mazda RX4 21 6 160 110 3.9 3.62 16.5 0 1 4
#> 2 Mazda RX… 21 6 160 110 3.9 3.88 17.0 0 1 4
#> 3 Datsun 7… 22.8 4 108 93 3.85 3.32 18.6 1 1 4
#> 4 Hornet 4… 21.4 6 258 110 3.08 4.22 19.4 1 0 3
#> 5 Hornet S… 18.7 8 360 175 3.15 4.44 17.0 0 0 3
#> 6 Valiant 18.1 6 225 105 2.76 4.46 20.2 1 0 3
#> # ℹ 2 more variables: carb <dbl>, .fitted <dbl>
newdata <- head(mtcars)
newdata$wt <- newdata$wt + 1
augment(n, newdata = newdata)
#> # A tibble: 6 × 13
#> .rownames mpg cyl disp hp drat wt qsec vs am gear
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Mazda RX4 21 6 160 110 3.9 3.62 16.5 0 1 4
#> 2 Mazda RX… 21 6 160 110 3.9 3.88 17.0 0 1 4
#> 3 Datsun 7… 22.8 4 108 93 3.85 3.32 18.6 1 1 4
#> 4 Hornet 4… 21.4 6 258 110 3.08 4.22 19.4 1 0 3
#> 5 Hornet S… 18.7 8 360 175 3.15 4.44 17.0 0 0 3
#> 6 Valiant 18.1 6 225 105 2.76 4.46 20.2 1 0 3
#> # ℹ 2 more variables: carb <dbl>, .fitted <dbl>
