Tidy summarizes information about the components of a model. A model component might be a single term in a regression, a single hypothesis, a cluster, or a class. Exactly what tidy considers to be a model component varies across models but is usually self-evident. If a model has several distinct types of components, you will need to specify which components to return.
Usage
# S3 method for class 'poLCA'
tidy(x, ...)Arguments
- x
A
poLCAobject returned frompoLCA::poLCA().- ...
Additional arguments. Not used. Needed to match generic signature only. Cautionary note: Misspelled arguments will be absorbed in
..., where they will be ignored. If the misspelled argument has a default value, the default value will be used. For example, if you passconf.lvel = 0.9, all computation will proceed usingconf.level = 0.95. Two exceptions here are:
See also
Other poLCA tidiers:
augment.poLCA(),
glance.poLCA()
Value
A tibble::tibble() with columns:
- class
The class under consideration.
- outcome
Outcome of manifest variable.
- std.error
The standard error of the regression term.
- variable
Manifest variable
- estimate
Estimated class-conditional response probability
Examples
# load libraries for models and data
library(poLCA)
library(dplyr)
# generate data
data(values)
f <- cbind(A, B, C, D) ~ 1
# fit model
M1 <- poLCA(f, values, nclass = 2, verbose = FALSE)
M1
#> Conditional item response (column) probabilities,
#> by outcome variable, for each class (row)
#>
#> $A
#> Pr(1) Pr(2)
#> class 1: 0.0068 0.9932
#> class 2: 0.2864 0.7136
#>
#> $B
#> Pr(1) Pr(2)
#> class 1: 0.0602 0.9398
#> class 2: 0.6704 0.3296
#>
#> $C
#> Pr(1) Pr(2)
#> class 1: 0.0735 0.9265
#> class 2: 0.6460 0.3540
#>
#> $D
#> Pr(1) Pr(2)
#> class 1: 0.2309 0.7691
#> class 2: 0.8676 0.1324
#>
#> Estimated class population shares
#> 0.2792 0.7208
#>
#> Predicted class memberships (by modal posterior prob.)
#> 0.3287 0.6713
#>
#> =========================================================
#> Fit for 2 latent classes:
#> =========================================================
#> number of observations: 216
#> number of estimated parameters: 9
#> residual degrees of freedom: 6
#> maximum log-likelihood: -504.4677
#>
#> AIC(2): 1026.935
#> BIC(2): 1057.313
#> G^2(2): 2.719922 (Likelihood ratio/deviance statistic)
#> X^2(2): 2.719764 (Chi-square goodness of fit)
#>
# summarize model fit with tidiers + visualization
tidy(M1)
#> # A tibble: 16 × 5
#> variable class outcome estimate std.error
#> <chr> <int> <dbl> <dbl> <dbl>
#> 1 A 1 1 0.00681 0.0254
#> 2 A 2 1 0.286 0.0393
#> 3 A 1 2 0.993 0.0254
#> 4 A 2 2 0.714 0.0393
#> 5 B 1 1 0.0602 0.0649
#> 6 B 2 1 0.670 0.0489
#> 7 B 1 2 0.940 0.0649
#> 8 B 2 2 0.330 0.0489
#> 9 C 1 1 0.0735 0.0642
#> 10 C 2 1 0.646 0.0482
#> 11 C 1 2 0.927 0.0642
#> 12 C 2 2 0.354 0.0482
#> 13 D 1 1 0.231 0.0929
#> 14 D 2 1 0.868 0.0379
#> 15 D 1 2 0.769 0.0929
#> 16 D 2 2 0.132 0.0379
augment(M1)
#> # A tibble: 216 × 7
#> A B C D X.Intercept. .class .probability
#> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl>
#> 1 2 2 2 2 1 1 0.959
#> 2 2 2 2 2 1 1 0.959
#> 3 2 2 2 2 1 1 0.959
#> 4 2 2 2 2 1 1 0.959
#> 5 2 2 2 2 1 1 0.959
#> 6 2 2 2 2 1 1 0.959
#> 7 2 2 2 2 1 1 0.959
#> 8 2 2 2 2 1 1 0.959
#> 9 2 2 2 2 1 1 0.959
#> 10 2 2 2 2 1 1 0.959
#> # ℹ 206 more rows
glance(M1)
#> # A tibble: 1 × 8
#> logLik AIC BIC g.squared chi.squared df df.residual nobs
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
#> 1 -504. 1027. 1057. 2.72 2.72 9 6 216
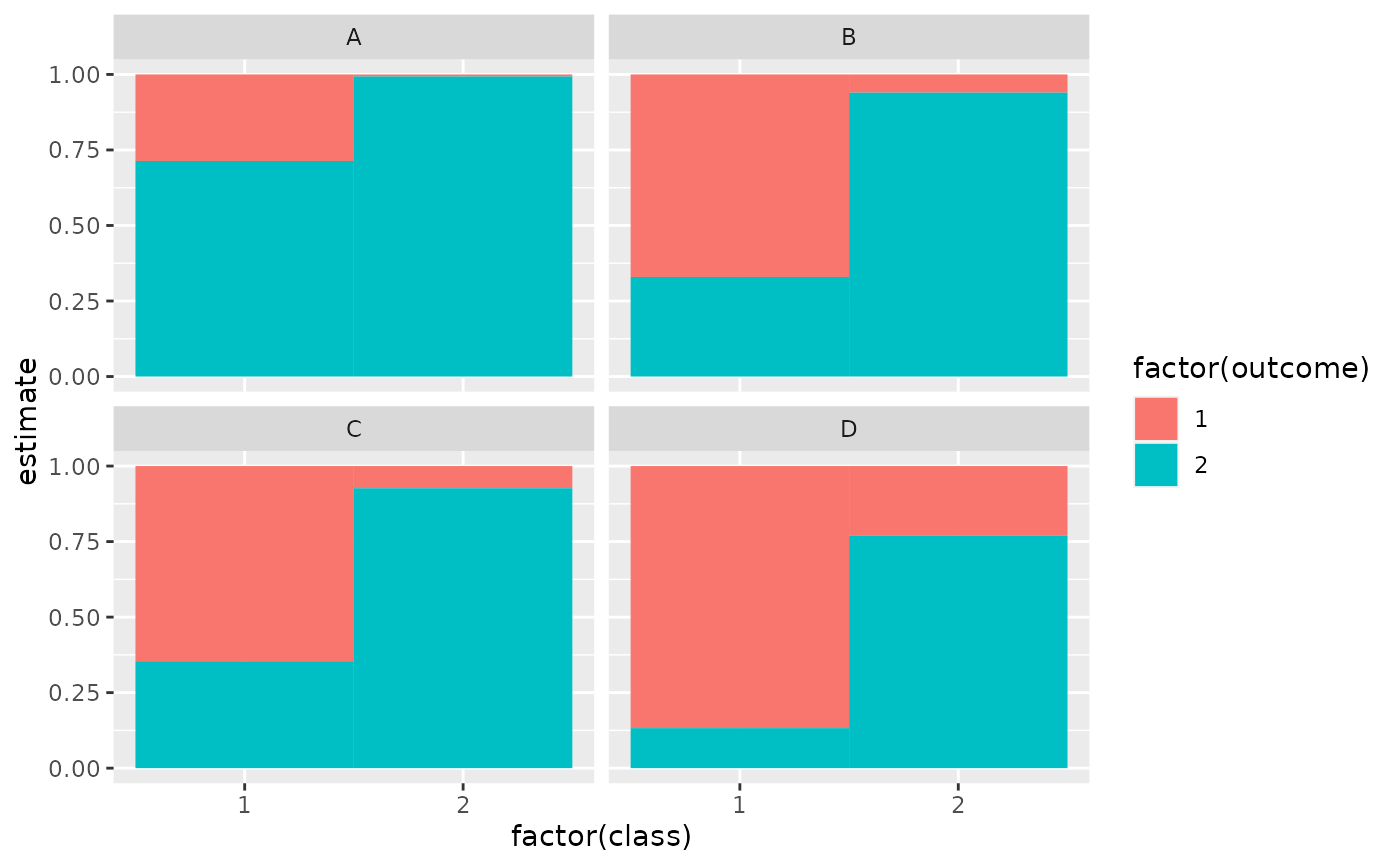
library(ggplot2)
ggplot(tidy(M1), aes(factor(class), estimate, fill = factor(outcome))) +
geom_bar(stat = "identity", width = 1) +
facet_wrap(~variable)
 # three-class model with a single covariate.
data(election)
f2a <- cbind(
MORALG, CARESG, KNOWG, LEADG, DISHONG, INTELG,
MORALB, CARESB, KNOWB, LEADB, DISHONB, INTELB
) ~ PARTY
nes2a <- poLCA(f2a, election, nclass = 3, nrep = 5, verbose = FALSE)
#> Error in eval(predvars, data, env): object 'MORALG' not found
td <- tidy(nes2a)
#> Error: object 'nes2a' not found
td
#> Error: object 'td' not found
ggplot(td, aes(outcome, estimate, color = factor(class), group = class)) +
geom_line() +
facet_wrap(~variable, nrow = 2) +
theme(axis.text.x = element_text(angle = 90, hjust = 1))
#> Error: object 'td' not found
au <- augment(nes2a)
#> Error: object 'nes2a' not found
au
#> Error: object 'au' not found
count(au, .class)
#> Error: object 'au' not found
# if the original data is provided, it leads to NAs in new columns
# for rows that weren't predicted
au2 <- augment(nes2a, data = election)
#> Error: object 'nes2a' not found
au2
#> Error: object 'au2' not found
dim(au2)
#> Error: object 'au2' not found
# three-class model with a single covariate.
data(election)
f2a <- cbind(
MORALG, CARESG, KNOWG, LEADG, DISHONG, INTELG,
MORALB, CARESB, KNOWB, LEADB, DISHONB, INTELB
) ~ PARTY
nes2a <- poLCA(f2a, election, nclass = 3, nrep = 5, verbose = FALSE)
#> Error in eval(predvars, data, env): object 'MORALG' not found
td <- tidy(nes2a)
#> Error: object 'nes2a' not found
td
#> Error: object 'td' not found
ggplot(td, aes(outcome, estimate, color = factor(class), group = class)) +
geom_line() +
facet_wrap(~variable, nrow = 2) +
theme(axis.text.x = element_text(angle = 90, hjust = 1))
#> Error: object 'td' not found
au <- augment(nes2a)
#> Error: object 'nes2a' not found
au
#> Error: object 'au' not found
count(au, .class)
#> Error: object 'au' not found
# if the original data is provided, it leads to NAs in new columns
# for rows that weren't predicted
au2 <- augment(nes2a, data = election)
#> Error: object 'nes2a' not found
au2
#> Error: object 'au2' not found
dim(au2)
#> Error: object 'au2' not found
